HIT Multimedia (Liu Peixiang, Zhang Delong / Text & Images) Recently, the research team led by Professor Haoyu Li from the Advanced Photonics and Imaging Center (IPIC) of the School of Instrumentation Science and Engineering, Harbin Institute of Technology (HIT), achieved a breakthrough in the field of biomedical super-resolution microscopy imaging technology. Addressing the current challenge of insufficient photon efficiency in live-cell super-resolution imaging, the team proposed an unsupervised learning-based self-inspired denoising method. Leveraging unsupervised deep learning, this method enhances photon efficiency by two orders of magnitude without the need for large training sets or high-signal-to-noise ratio (SNR) ground truth images, enabling gentle, long-term in vivo imaging under low-illumination conditions. For a long time, existing microscopy technologies have been unable to balance requirements such as low phototoxicity, high spatial resolution, and high temporal resolution. This technology breaks this bottleneck by denoising with only a single noisy frame, successfully capturing a 3-hour-long five-dimensional (xyz-color-time) mitosis process with a resolution of 90 nm on a spinning disk confocal super-resolution microscopy system. With broad applicability across various super-resolution systems, this technology provides a new scientific imaging tool for studying organelle interaction mechanisms and other biomedical research at the super-resolution scale.
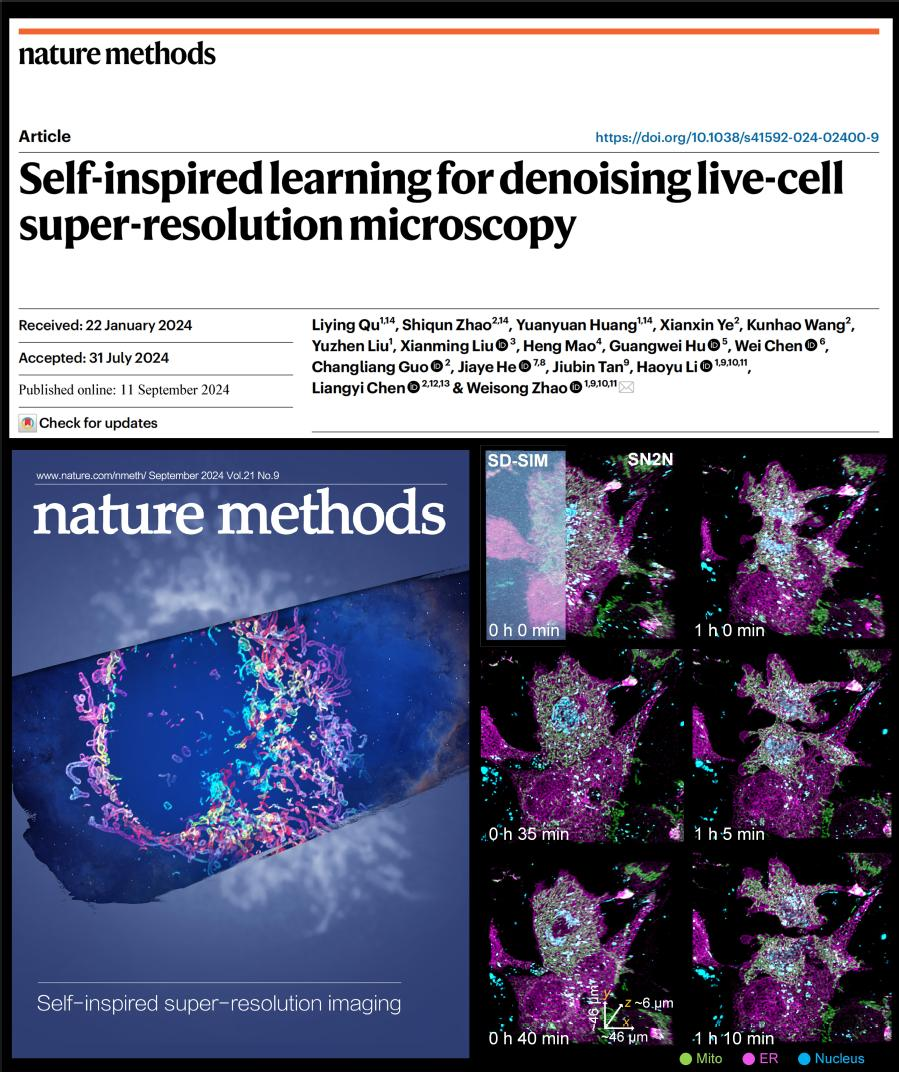
On September 11, the research results, titled Self-inspired learning for denoising live-cell super-resolution microscopy, were published online as a full-length article in the international authoritative journal《Nature Methods》(2023 Impact Factor: 49.0).
The development goal of live-cell super-resolution fluorescence microscopy is to maintain sufficient spatiotemporal resolution under physiologically friendly imaging conditions. However, improving spatial resolution typically requires increasing illumination intensity or extending exposure time, while matching temporal resolution to prevent motion artifacts—making the enhancement of photon efficiency crucial in live-cell super-resolution imaging. Deep neural networks use supervised learning to fit the mapping between noisy and clean images, which can significantly improve photon efficiency. Yet, they require collecting a large number of paired clean images, making them difficult to apply to live cells. Unsupervised learning denoising methods offer an alternative approach to enhance photon efficiency in super-resolution imaging but still require a large number of noisy images for training, resulting in limited denoising effects and low data efficiency. Therefore, developing an unsupervised learning denoising method to improve the photon efficiency of super-resolution microscopy systems and achieve long-term live-cell imaging at the super-resolution scale with limited data and without the need for noisy image pairs remains a challenge in the field.
To address these issues, Professor Haoyu Li's team proposed a self-inspired learning super-resolution denoising method (Self-inspired Noise2Noise, SN2N for short). SN2N leverages the spatial sampling redundancy of super-resolution systems to design a self-supervised data generation strategy, producing the required noisy image pairs from a single image as the dataset. It also develops a self-constrained learning strategy to further improve denoising performance and data efficiency, bringing denoising effects close to those of supervised learning. Ultimately, SN2N can enhance the photon flux of super-resolution microscopy systems by more than two orders of magnitude. Additionally, SN2N can be integrated with various commonly used optical super-resolution microscopy technologies, serving as an easy-to-use photon flux enhancement tool. The method also includes a high-precision segmentation solution for organelle structures and provides intelligent pre-analysis functions, promising to facilitate downstream high-throughput biological information classification and high-precision intelligent segmentation and tracking of organelles.
Four-Color Live-Cell Super-Resolution Imaging—Revealing Organelle Interaction Phenomena
The research team further combined the SN2N method with deconvolution technology (RL-SN2N) to improve resolution and applied it to a Spinning Disk Confocal Structured Illumination Microscopy (SD-SIM) system, achieving clear four-color live-cell imaging (Image 1). SN2N enables the visualization of fast and complex organelle dynamic processes. By detailed recording the trajectories and spatial distributions of different organelles, it tracks interaction events between multiple organelles, facilitating in-depth exploration of their synergistic effects and interaction relationships.
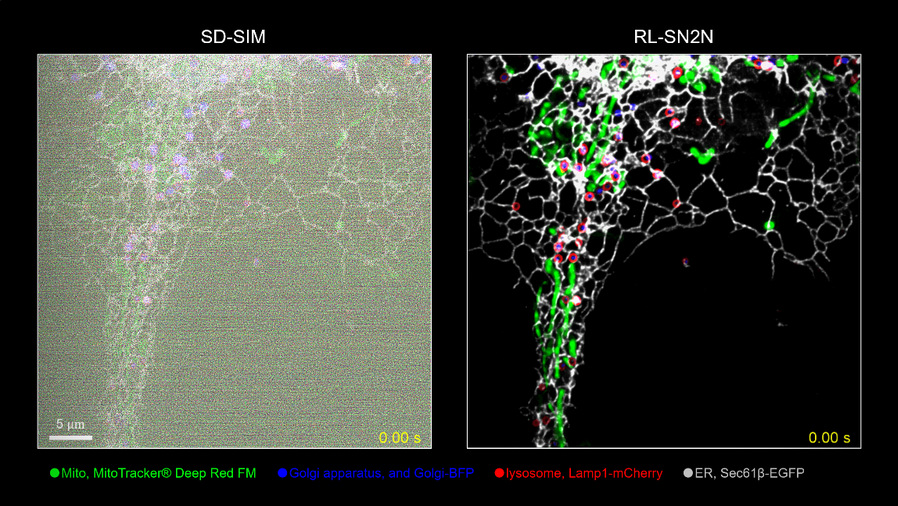
Image 1: Four-color live-cell imaging results of SN2N on the SD-SIM system: mitochondria (green), endoplasmic reticulum (gray), lysosomes (red), and Golgi apparatus (blue).
Five-Dimensional Long-Term Live-Cell Super-Resolution Imaging—Completely Recording the Mitosis Process
The research team also used the developed method to successfully achieve five-dimensional (xyz-color-time) long-term live-cell super-resolution imaging with a resolution of up to 90 nm on the SD-SIM system. This technology reveals the dynamic omics interactions between the endoplasmic reticulum (magenta), mitochondria (green), and cell nucleus (cyan) throughout the entire cell mitosis process, with a monitoring time exceeding 3 hours. It provides important experimental evidence for biologists to further explore the functions and interaction relationships of organelles (Image 2).
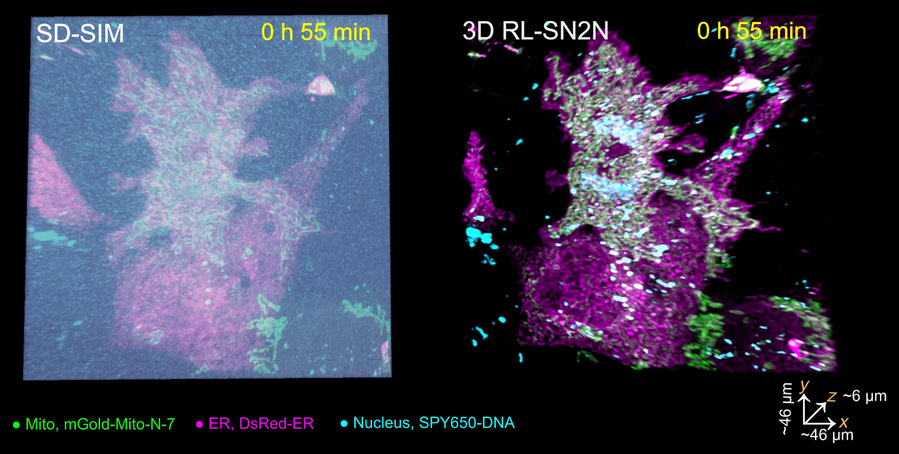
Image 2: SN2N enables five-dimensional long-term live-cell imaging, recording the complete mitosis process.
Metaphorically, the SN2N technology is like a magic mirror that transforms blurry images into clear pictures through self-reflection and self-inspiration, gradually revealing elusive biological information. It acts as an excellent decoder, uncovering the secrets of the biological world hidden in the fog. By cleverly leveraging the physical characteristics of super-resolution microscopy systems without the need for a large number of matched images, SN2N makes gentle, long-term super-resolution imaging of live cells a reality. Not only does it break the bottlenecks of traditional super-resolution microscopy in terms of photon efficiency and data requirements, but it can also be applied to various existing super-resolution microscopy systems. With SN2N, biologists can clearly observe complex dynamic changes inside cells at long-term, high-resolution scales, thereby interpreting cellular functions and mechanisms more accurately. The application of this technology provides a new perspective for in-depth exploration of intracellular structures and a powerful tool for biomedical research and disease mechanism analysis.
This paper is the first《Nature Methods》article with HIT as the first completing unit. Qu Liying, a doctoral student from the School of Instrumentation Science and Engineering of HIT, is the first author of the paper. Associate Researcher Zhao Shiqun from Peking University and Huang Yuanyuan, a doctoral student from the School of Instrumentation Science and Engineering of HIT, are co-first authors. Professor Zhao Weisong from the School of Instrumentation Science and Engineering of HIT is the corresponding author, and Professor Haoyu Li and Academician Tan Jiubin are co-authors. This work was supported by the Key Research and Development Program of the Ministry of Science and Technology and the National Natural Science Foundation of China.
Paper link: https://www.nature.com/articles/s41592-024-02400-9

